The Texas A&M Drug Discovery and Development Resource Center (3DRC)
Is a Texas A&M-approved and regulated BSL2 facility that occupies approximately 7000 sq. ft. in the TMC3 Helix Park Collaborative Research Building, Texas Medical Center, Houston, Texas. It supports all stages of in silico and in vitro preclinical drug discovery and development. It is well-equipped to perform virtual screening, medicinal chemistry, high-throughput screening, and data analysis in this proposal. The core provides industry-standard computational resources, synthetic chemistry, liquid handling, and high-throughput endpoint detection capabilities for scientists conducting chemical, chemical-genomic, and drug discovery research. It can also support biochemical, cell-based, and foundational organism library screens. In addition to traditional high-throughput screening, our facility supports the development of specialized capabilities for lower-throughput drug discovery and development needs, including optical metabolic imaging, microelectrode arrays, and impedance systems for discovery and toxicity testing, as well as spectral flow for high-complexity biomarker development and testing.
The Gulf Coast Consortium for Innovative Drug Discovery and Development
The Consortium for Innovative Drug Discovery and Development (GCC IDDD) is a research consortium formed in 2003 that is focused on providing support for Houston/Galveston scientists in advancing their therapeutics discoveries through development to the clinic. Consortium support includes collaborative networking and joint funding opportunities, shared core resources, and educational programs. 3DRC is a designated core resource facility available through the IDDD.
The CPRIT Cancer Therapeutics Training Program
The Cancer Therapeutics Training Program (CTTP) is a multi-institutional post-doctoral training program funded by the CPRIT and is designed to prepare post-doctoral trainees for future careers in academic and/or commercial cancer therapeutics research and development. The goal of the program is to recruit and train scientists equipped with the essential skills and knowledge necessary to translate basic cancer research discoveries into commercially viable cancer therapeutics. The 3DRC accepts CTTP-fellows for core rotations and other training opportunities.
Computing Resources
DDRC-Dedicated Computing
 3DRC maintains a network of computational workstations and servers collectively functioning as a distributed grid computing cluster. In total, this network provides more than 400 CPU cores and over 4 TB of high-speed RAM, providing ample computational power for a wide range of image analysis and conventional data modeling techniques. More specifically the dedicated 3DRC cluster includes multiple specialized high-capacity GPU compute nodes with one node four NVIDIA A6000 Ada 48Gb GPUS, seven nodes containing dual NVIDIA RTX A6000/48Gb with SLI, one node with a single NVIDIA RTX A6000/48Gb and one with a NVIDIA Tesla V100 (32GB), collectively providing ample resources for training and deploying deep learning driven workflows. The core laboratory also maintains a centralized image and data repository of more than 700TB with a redundant mirrored backup system maintained by the Texas A&M Office of Information Technology in College Station, TX. Multiple software are used to support drug design and cheminformatics, including PharmacoNet, DiffDock, GNINA, Vina, QuickVina, ChimeraX, BioVia Draw, KNIME, Boltz-2, and Pipeline Pilot. Multiple generative open-source ligand design software has been implemented, including Dragonfly, Polygon, Synthemol, SPMM, and DrugGPT. New methods are continually assessed and incorporated as needed. Data analysis from in vitro biochemical and cell-based assays is performed using a combination of Pipeline Pilot from Biovia, Microsoft Office 365, R, Python, and GraphPad Prism. High-content image analysis algorithms are developed and executed using the IN-Cell Developer, MetaXpress Analysis (Molecular Devices), or Pipeline Pilot (Biovia). Deep learning models are developed and trained in either Pipeline Pilot (Bio-via), or in Python/Pytorch. Virtualization software includes WSL2 and Docker. Operating systems include Windows 11, Windows Server 2022, Ubuntu 22.04, and Red Hat Enterprise Linux. Additional software includes Microsoft 365 Suite, Adobe CC Suite, Graphpad Prism, FlowJo. Additional computational resources are made available to all Texas A&M faculty and staff through the HPRC resource center (https://hprc.tamu.edu/ ) and cloud computing resources.
3DRC maintains a network of computational workstations and servers collectively functioning as a distributed grid computing cluster. In total, this network provides more than 400 CPU cores and over 4 TB of high-speed RAM, providing ample computational power for a wide range of image analysis and conventional data modeling techniques. More specifically the dedicated 3DRC cluster includes multiple specialized high-capacity GPU compute nodes with one node four NVIDIA A6000 Ada 48Gb GPUS, seven nodes containing dual NVIDIA RTX A6000/48Gb with SLI, one node with a single NVIDIA RTX A6000/48Gb and one with a NVIDIA Tesla V100 (32GB), collectively providing ample resources for training and deploying deep learning driven workflows. The core laboratory also maintains a centralized image and data repository of more than 700TB with a redundant mirrored backup system maintained by the Texas A&M Office of Information Technology in College Station, TX. Multiple software are used to support drug design and cheminformatics, including PharmacoNet, DiffDock, GNINA, Vina, QuickVina, ChimeraX, BioVia Draw, KNIME, Boltz-2, and Pipeline Pilot. Multiple generative open-source ligand design software has been implemented, including Dragonfly, Polygon, Synthemol, SPMM, and DrugGPT. New methods are continually assessed and incorporated as needed. Data analysis from in vitro biochemical and cell-based assays is performed using a combination of Pipeline Pilot from Biovia, Microsoft Office 365, R, Python, and GraphPad Prism. High-content image analysis algorithms are developed and executed using the IN-Cell Developer, MetaXpress Analysis (Molecular Devices), or Pipeline Pilot (Biovia). Deep learning models are developed and trained in either Pipeline Pilot (Bio-via), or in Python/Pytorch. Virtualization software includes WSL2 and Docker. Operating systems include Windows 11, Windows Server 2022, Ubuntu 22.04, and Red Hat Enterprise Linux. Additional software includes Microsoft 365 Suite, Adobe CC Suite, Graphpad Prism, FlowJo. Additional computational resources are made available to all Texas A&M faculty and staff through the HPRC resource center (https://hprc.tamu.edu/ ) and cloud computing resources.
TAMU-HPRC Shared Computing
Texas A&M University’s High Performance Research Computing (HPRC) provides advanced computing resources to support the research and instructional activities of its academic community. All Texas A&M students, faculty, and staff engaged in academic research or teaching are eligible to access HPRC resources at no cost. External collaborators may also gain access through sponsorship by a Texas A&M principal investigator (PI).HPRC operates several high-performance computing clusters designed to meet diverse research needs:
Grace: The flagship heterogeneous HPC cluster, Grace comprises 800 standard compute nodes, 100 NVIDIA A100 GPU nodes, 17 nodes with T4 or RTX6000 GPUs, and 8 large-memory nodes with 3 TB RAM. It utilizes Mellanox HDR100 InfiniBand interconnects and offers over 5 PB of high-performance storage via a Lustre parallel file system.
ACES: ACES is a Dell x86 HPC cluster featuring 130 nodes totaling 11,888 cores, including Intel Sapphire Rapids, Ice Lake, and AMD Rome processors. It incorporates a variety of accelerators such as 30 NVIDIA H100 GPUs, 4 NVIDIA A30 GPUs, Graphcore IPUs, NEC Vector Engines, and Intel FPGAs. The system employs NVIDIA Mellanox NDR200 InfiniBand interconnects and provides 2.3 PB of storage through a DDN Lustre appliance .
FASTER: FASTER is a composable high-performance data-analysis and computing instrument funded by the NSF MRI program. It consists of 180 compute nodes with Intel Xeon 8352Y Ice Lake processors, each with 256 GB RAM. The system features a pool of GPUs, including 200 NVIDIA T4, 40 A100, 8 A10, 4 A30, and 8 A40 GPUs, which can be dynamically allocated to compute nodes via Liqid PCIe Gen4 fabrics. FASTER utilizes Mellanox HDR100 InfiniBand interconnects and offers 5 PB of usable storage with a Lustre file system.
Terra: Terra is a Lenovo x86 HPC cluster comprising 320 compute nodes with a total of 9,632 cores. The cluster includes 256 nodes with 64 GB RAM, 48 GPU nodes equipped with NVIDIA K80 GPUs and 128 GB RAM, and 16 Intel Knights Landing (KNL) nodes with 96 GB RAM. Terra employs Intel Omni-Path Architecture (OPA) interconnects and provides 7.4 PB of raw storage capacity.
These resources empower the Texas A&M research community to perform cutting-edge computational and data-intensive research across various scientific and engineering disciplines.
Chemistry Resources
Synthetic Chemistry
The newly established synthetic chemistry laboratory is equipped with state-of-the-art instrumentation that enables rapid compound synthesis, purification, and characterization essential for advanced research projects. The laboratory features six 8-foot chemical fume hoods with appropriate glassware and scaffolding to ensuring safe handling of chemicals during synthesis. For purification processes, the laboratory houses a Buchi Prep C-830 preparative HPLC system capable of chiral separations using chiral columns, and a Biotage Isolera One flash chromatography instrument for automated purification of reaction mixtures. Solvent removal and sample concentration are efficiently managed using three Buchi Rotavap R-300 rotary evaporators with F-305 recirculating chillers. A Labconco FreeZone 2.5 L -50°C benchtop lyophilizer provides freeze-drying capabilities for peptides and small molecules. Analytical characterization is supported by a Shimadzu LC-MS system with a mass spectrometry detector, enabling compound characterization and real-time reaction monitoring. A Bruker Fourier 80 benchtop NMR spectrometer with 1H and 13C probes offers capabilities for both one-dimensional and two-dimensional NMR studies, essential for structural elucidation. The laboratory also includes a Mettler Toledo Karl Fischer titrator for accurate moisture analysis and two Mettler Toledo analytical balances for precise weighing of reagents. Additional equipment includes two biosafety cabinets for handling sensitive materials and ensuring a contamination-free environment, as well as a VWR ultrasonic sonicator for efficient sample preparation. An automated peptide synthesizer facilitates efficient and precise peptide assembly, while parallel block reactors and curated structure-activity relationship (SAR) kits from Sigma-Aldrich allow for expedited synthesis and diversification of compounds, accelerating SAR studies and the development of in silico hits. This comprehensive suite of advanced instrumentation positions the laboratory to effectively support high-level synthetic chemistry research, promoting innovation and accelerating the development of novel compounds.
Drug Libraries
A detailed description of drug libraries can be found here. A complete list of all (active and previously offered) drug libraries can be viewed and downloaded here
| Library Name | Approved | Mechanistic | Status | Link |
|---|---|---|---|---|
| Broad 2021 | Yes | Yes | Active | SDF / XLSX |
| MCE FDA and Pharmacopia (2024) | Yes | Yes | Active | SDF / XLSX |
| MCE Epigenetics | Yes | Yes | Active | SDF / XLSX |
| SGC Epigenetics | Yes | Yes | Active | SDF / XLSX |
| CCI 2022 | Yes | Yes | Active | SDF / XLSX |
| NCI AOD 11 | Yes | Yes | Active | SDF / XLSX |
| TargetMol L1000 Approved Drug Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L1200 Epigenetics Compound Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L1300 PI3K AKT and mTOR Compound Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L1400 MAPK Inhibitor Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L2200 Tyrosine Kinase Inhibitor Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L2400 Endocrinology Hormones Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L2600 Neuronal Signaling Compound Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L2900 Oxidation and Reduction Compound Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L3200 Autophagy Compound Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L3700 JAK STAT Compound Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L3900 DNA Damage & Repair Compound Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L4000 Bioactive Compound Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L4300 Wnt Hedgehog and Notch Compound Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L5100 Fluorochemical Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L5200 Metabolic Disease Compound Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L5300 Mitochondrial Targeting Compound Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L6000 Natural Compound Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L8000 Stem Cell Differentiation Compound Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L8100 Cell Cycle Related Compound Library | Yes | Yes | Active | SDF / XLSX |
| TargetMol L9000 Apoptosis Compound Library | Yes | Yes | Active | SDF / XLSX |
| UTKinase V5 | Yes | Yes | Active | SDF / XLSX |
Screening Resources
Automated High-Throughput Screening Platforms
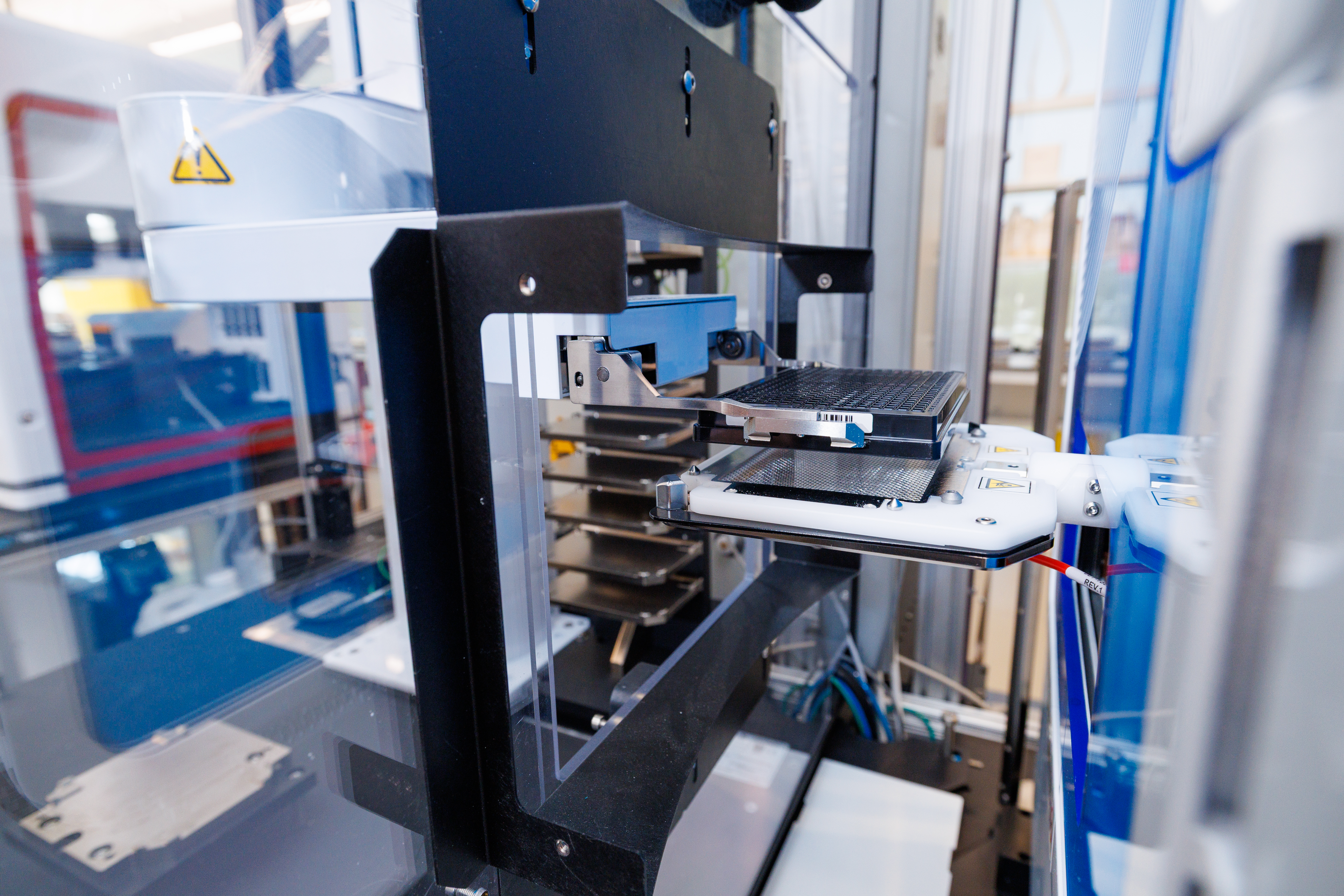 The 3DRC maintains and operates two major automated platforms for setting up in vitro high-throughput screening (HTS) assays, plus a fully integrated wash station and a suite of smaller walk-up systems for semi-automated workflows:
The 3DRC maintains and operates two major automated platforms for setting up in vitro high-throughput screening (HTS) assays, plus a fully integrated wash station and a suite of smaller walk-up systems for semi-automated workflows:
- Automated Labcyte Echo 655: A “no-touch” acoustic dispenser that uses focused sound energy to transfer compounds from source plates to assay plates in sub-microliter volumes, minimizing cross-contamination and maximizing precision. An integrated Access robotic arm handles plates between the Echo, a Thermo Cytomat 6000A automated cell-culture incubator (189-plate capacity), and a Thermo Multidrop Combi reagent dispenser, fully automating the assay-setup workflow.
- Biomek i7 liquid-handling platform: Configured with a 96-channel pipetting head, an independent Span-8 arm, 100-nL pintools, and a rotating gripper, this system delivers both high precision and flexible deck access. It supports multiple plate formats and a wide range of HTS protocols—from reagent titrations to complex multi-step assays.
- Automated wash and reagent-addition station: Combines a Tecan HydroSpeed microplate washer with a Thermo Orbitor plate stacker (40-plate capacity) and an integrated Thermo Multidrop Combi dispenser. This station streamlines wash cycles, reagent additions, and plate storage to keep throughput high and hands-on time low.
- Standalone “walk-up” and semi-automated systems: Four Thermo Multidrop Combi dispensers and two Formulatrix Mantis liquid handlers are available for lower-throughput or user-driven assays. These bench-level platforms provide rapid turnaround for pilot screens, assay development, and follow-up experiments.
Automated Multi-modal Plate Reader
The DDRC has a Synergy Neo2, manufactured by Agilent BioTek, which is a high-performance, hybrid multi-mode microplate reader engineered for high-throughput screening (HTS) applications. This system is integrated with the BioStack Neo plate stacker, delivering fully automated, walk-away operation for a wide range of assay formats, including biochemical, cell-based, and kinetic assays. The Synergy Neo2 employs patented Hybrid Technology, combining independent filter- and quadruple monochromator-based optical paths for optimal performance across detection modes such as UV-Vis absorbance, fluorescence intensity, luminescence, fluorescence polarization, time-resolved fluorescence (TRF), TR-FRET, BRET, and AlphaScreen®/AlphaLISA®. The reader features continuously variable bandwidths (3–50 nm in 1 nm increments), up to four photomultiplier tubes (PMTs) for ultra-fast measurements, and environmental controls including incubation to 70°C, CO₂/O₂ regulation, and programmable shaking, making it suitable for live-cell assays. The BioStack Neo plate stacker complements the reader by automating plate handling, with a 50-plate capacity, rapid plate exchange, and automated de-lidding features to enhance throughput and consistency, particularly for sensitive cell-based assays.
Surface Plasmon Resonance (SPR)
3DRC has dedicated access to a Biacore T200 SPR biosensor. This is a high-performance system for real-time analysis of biomolecular interactions. It supports up to four flow cells in series, with paired reference subtraction to deliver precise affinity and kinetic data across a broad range of interactant sizes. It is further integrated with an autosampler to accommodate 96- or 384-well microplates, enabling unattended injection of hundreds of samples for high-throughput screening of binding kinetics, concentration determination, and specificity assays. Low-dead-volume fluidics, temperature control (4 – 40 °C), and the BioCore V4 chip ensure ultra-low noise, picomolar sensitivity, and reproducible on-/off-rate measurements; even for interactions at the extremes of the kinetic scale. The user-friendly Biacore T200 Control and Evaluation software feature guided wizards, Method Builder for custom assay design, and advanced data-analysis modules (single-cycle kinetics, heterogeneous ligand models, screening evaluation) to streamline setup, execution, and interpretation of complex
Isothermal Calorimetry (ITC)
3DRC also offers ITC using its Malvern Microcal PEAQ-ITC for direct, label-free measurement of the thermodynamic forces driving molecular interactions. ITC determines key binding parameters, including affinity (KD), stoichiometry (n), enthalpy (ΔH), and entropy (ΔS) in a single experiment, without the need for immobilization, labeling, or surface attachment. Operating under near-physiological conditions, ITC is ideal for studying protein–ligand, protein–protein, and nucleic acid interactions. Unlike SPR, which excels at kinetic profiling, ITC uniquely reveals the complete thermodynamic fingerprint of a binding event, quantifying the energetic forces that drive molecular recognition, such as hydrogen bonding, hydrophobic interactions, and conformational changes.
Automated Flow Cytometry
 For multiplex high-throughput screening of nonadherent cell-based models, the core uses an automated flow cytometry platform. The heart of the platform is a 5-laser, 30-detector BioRad ZE5 high-throughput flow cytometer capable of running uninterrupted for up to 20 hours. The flow cytometer is integrated with an integrated robotics workstation that provides a structure to hold all equipment in a fixed orientation around an articulating Universal Robotics 6-axis UR10 robotic plate manipulator. The platform also includes integrated peripheral devices to automate most experimental procedures needed for routine flow analysis. This equipment consists of an automated cell culture incubator, a refrigerated incubator to hold plates at 4ºC for extended periods, an on-deck automated centrifuge, a deep-well plate washer, a multi-reagent bulk liquid dispenser and plate shaker, a Formulatrix Mantis automated microfluidic dispenser, a microplate reading bar code scanner, and a room temperature storage and de-lidding device. All of the scheduling and integration is handled using Hudson Robotic’s Softlinx software.
For multiplex high-throughput screening of nonadherent cell-based models, the core uses an automated flow cytometry platform. The heart of the platform is a 5-laser, 30-detector BioRad ZE5 high-throughput flow cytometer capable of running uninterrupted for up to 20 hours. The flow cytometer is integrated with an integrated robotics workstation that provides a structure to hold all equipment in a fixed orientation around an articulating Universal Robotics 6-axis UR10 robotic plate manipulator. The platform also includes integrated peripheral devices to automate most experimental procedures needed for routine flow analysis. This equipment consists of an automated cell culture incubator, a refrigerated incubator to hold plates at 4ºC for extended periods, an on-deck automated centrifuge, a deep-well plate washer, a multi-reagent bulk liquid dispenser and plate shaker, a Formulatrix Mantis automated microfluidic dispenser, a microplate reading bar code scanner, and a room temperature storage and de-lidding device. All of the scheduling and integration is handled using Hudson Robotic’s Softlinx software.
Automated High-Throughput Microscopy Platforms

Both ImageXpress systems offer flexibility in balancing imaging speed and quality. Software-controlled objectives allow users to switch from low magnification (2×/0.10 NA) for ultra-high throughput—up to approximately 200,000 images/day—to high magnification (40×/0.95 NA) for high-content imaging. The software also facilitates seamless switching between confocal and wide-field imaging modes without manual reconfiguration, enabling sequential imaging of the same plate in both modes. The systems support multiplex fluorescence assays with simultaneous detection of up to five fluorophores and are compatible with Förster resonance energy transfer (FRET)-based reporters. Both platforms are integrated with automated cell culture incubators, bulk liquid dispensers for reagent addition, and robotic arms for plate handling and peripheral devices.
Automated Optical Metabolic Imaging
Changes in cellular metabolism have long been recognized as a fundamental feature, and are now considered a hallmark, of cancer. The 3DRC employs an advanced two-photon fluorescent lifetime microscopy metabolic imaging method to significantly enhance our understanding of how specific microenvironmental factors influence the regulation of cancer cell metabolism and drug responsiveness at a single-cell level. This platform utilizes a non-invasive, quantitative measure of the intrinsic fluorescence of reduced nicotinamide adenine dinucleotide (NADH), a natural biomarker involved in various critical cellular processes, including mitochondrial function, energy metabolism, calcium homeostasis, gene expression, oxidative stress, aging, and apoptosis. This platform also measures the autofluorescence of flavin adenine dinucleotide (FAD) that is localized in the mitochondria and is involved in the TCA cycle and complex II in the electron transport chain. The 3DRC platform is from Intelligent Imaging Innovations (3i) and employs femtosecond laser pulses (680-1080 nm, 140 fs, 76 MHz) generated using Chameleon Ultra II Ti:Sapphire Laser system (Thorlabs Inc., Newton, NJ). The laser pulses were steered toward dual-channel time-correlated single photon counting (TCSPC) electronics and an inverted microscope (Zeiss Axio Observer Z1 fully motorized inverted microscope) fitted with environmental controls for temperature, humidity, and CO2. The platform has a motorized X,Y stage with 0.1µm linear encoders to support the use of either slides or multi-well plates (96-384well/plate). This platform is equipped for both live and fixed cell widefield epifluorescence applications, transmitted illumination, and multi-photon techniques.
Micro-electrode array
Axion Maestro Pro electrophysiology and impedance platform is a label-free, high-throughput multimodal bioelectronic assay system combining multi-well microelectrode array (MEA) recording (6–96 wells) and impedance-based assays (96–384 wells), with the BioCore V4 processing chip for ultra-low noise and high signal fidelity. This platform supports the majority of MLOTS standardized screening workflows, which include neurotoxicity (network activity metrics, burst and synchrony analysis) and cardiotoxicity (field potential duration, beat detection) with real-time monitoring of electrophysiological endpoints using iPSC and mouse cell models. Impedance measurements enable continuous tracking of cell proliferation, barrier function, co-culture dynamics, and long-term cell growth. These are critical dynamic endpoints for chronic toxicity and co-culture analysis assays. Integrated environmental control (heating, CO₂, humidity), barcode tracking, and AxIS software streamline assay setup, automated data acquisition, and analysis for robust throughput screening.
TAMU-IBT Advanced Microscopy Core
Several conventional high-content imaging systems are available to 3DRC through the TAMU-IBT Advanced Microscopy Core. For high content 3-D imaging or where oil-immersion is preferred, the 3DRC uses a Nikon W1 Yokagawa spinning disk confocal or a General Electric Healthcare DeltaVision Elite deconvolution microscope. Both of these platforms feature climate-controlled (temperature, humidity, CO2) automated stages, which provide a large range of live cell capabilities. Both microscopes have multidimensional acquisition (x,y,z, wavelength, time), up to 5 channels (fluorescence and brightfield/DIC), FRET, multi-point acquisition, large area acquisition (tiling), dual Autofocus (Hardware & contrast based), fast acquisition mode and 4x,10x,20x,40x air, 40x Oil, 60x oil, 100x oil objectives.